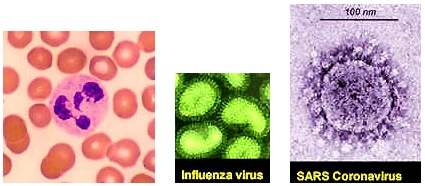
1 micrometer (micron) = 1 / millionth of a meter (39.37 inches) 1 nanometer = 1 / billionth of a meter
August 21, 2004 West LaFayette, Indiana - When you enter the world of bio-nanotechnology, you are so small that if one nanometer were the size of a single pea, a regular meter of 39.37 inches would be the size of the Earth! In that nano world, virus sizes vary from 18 to 300 nanometers.
Click here to subscribe and get instant access to read this report.
Click here to check your existing subscription status.
Existing members, login below:
© 1998 - 2024 by Linda Moulton Howe.
All Rights Reserved.

